✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
DNeasy Blood & Tissue Kit (50)
Cat. No. / ID: 69504
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Standardized method for a variety of sample types
- High yields even from specialized samples
- High-quality DNA
- Optimized protocols for a range of starting materials
- Spin-column and 96-well high-throughput formats
- Automate workflows on the QIAcube Connect
Product Details
DNeasy Blood & Tissue Kits provide fast, easy silica-based DNA extraction without phenol or chloroform in convenient spin-column and 96-well-plate formats. Most samples can be directly lysed with proteinase K, eliminating the need for mechanical disruption and reducing hands-on time. Optimized protocols for specific sample types provide reproducible extraction of high-quality DNA for life science, genotyping, and veterinary pathogen research applications. Extracting DNA using the DNeasy Blood & Tissue Kit can be automated on the QIAcube Connect.
DNeasy Blood & Tissue standard protocols can also be executed using the TRACKMAN Connected system, paired with PIPETMAN M Connected pipettes, both from Gilson. The TRACKMAN Connected system guides researchers through the DNeasy Blood & Tissue protocols while automatically adjusting the Bluetooth-enabled PIPETMAN M Connected pipette settings. Each step of the protocol execution is recorded to accelerate reporting by generating a comprehensive run report. Download more information.
In partnership with My Green Lab, we've also assessed the environmental impact of the DNeasy Blood & Tissue Kit (250). My Green Lab ACT environmental impact factor labels are designed to evaluate and score products on several sustainability criteria, including:
- Manufacturing
- Responsible chemical management
- Sustainable content within products and packaging materials
- Disposal of the packaging at the end of life
Products are scored from 1 to 10 except for energy and water consumption, which are scored as 1 point per kWh or gallon, respectively. A low score means a lower environmental impact (see figures "DNeasy Blood & Tissue Kit environmental impact factor label US, EU and UK").
For an eco-friendlier alternative to this kit, see our QIAwave DNA Blood & Tissue Kit.
See figures
Performance
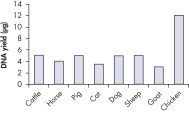
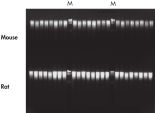
The efficient DNeasy Blood & Tissue procedure enables high yields of total DNA from animal blood and tissue samples (see table Typical DNA yields from animal tissues using DNeasy Blood & Tissue Kits and figure DNA yields). Optimized protocols ensure high yields from nonstandard samples, such as animal hair (see Genotyping of horses), as well as cultured cells, fixed tissues, or Gram-positive and -negative bacteria. Specialized online protocols are available for DNA purification from yeast, insects, hair, and other sample types.
| Source | Amount | DNA (µg) |
| Mammalian blood | 100 µl | 3–6 |
| Bird blood | 5 µl | 9–40 |
| HeLa cells | 2 x 106 | 15–25 |
| Liver | 25 mg | 10–30 |
| Brain | 25 mg | 15–30 |
| Kidney | 25 mg | 15–30 |
| Spleen | 10 mg | 5–30 |
| Mouse tail | 1.2 cm (tip) | 10–25 |
| Rat tail | 0.6 cm (tip) | 20–40 |
| Pig ear | 25 mg | 10–30 |
| Horse hair | 10 hairs | 2–4 |
| Fish fin | 20 mg | 10–20 |
| Fish spawn (mackerel) | 10 mg | 5–10 |
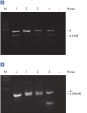
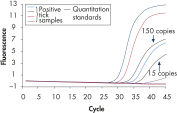
DNeasy Blood & Tissue Kits simplify purification of DNA from a wide range of sample types, including animal species commonly encountered in life science, veterinary, and genotyping applications (see High-quality DNA). Purified DNA is free from PCR inhibitors, enabling sensitive detection in standard, multiplex (see Efficient 16plex PCR), and real-time PCR (see Real-time PCR). DNeasy Blood & Tissue Kits provide reliable results — from laboratory analysis of transgenic mice (see High-throughput purification) to livestock breeding programs (see Genotyping of pigs) and pedigree genotyping. Routine testing applications can be easily scaled up using the DNeasy 96 Blood & Tissue Kit (see High-throughput purification).
See figures
Principle
DNeasy Blood & Tissue Kits are designed for rapid purification of total DNA (e.g., genomic, mitochondrial, and pathogen) from a variety of sample sources including fresh or frozen animal tissues and cells, blood, or bacteria.
The DNeasy membrane combines the binding properties of a silica-based membrane with simple microspin technology or with the QIAGEN 96-Well-Plate Centrifugation System. DNA adsorbs to the DNeasy membrane in the presence of high concentrations of chaotropic salt, which remove water from hydrated molecules in solution. Buffer conditions in DNeasy Blood & Tissue procedures are designed to enable specific adsorption of DNA to the silica membrane and optimal removal of contaminants and enzyme inhibitors.
Purification requires no phenol or chloroform extraction or alcohol precipitation, and involves minimal handling. This makes DNeasy Blood & Tissue Kits highly suited for simultaneous processing of multiple samples. For higher-throughput applications, the DNeasy 96 Blood & Tissue Kit enables simultaneous processing of 96 or 192 samples.
Procedure
Reliable silica-membrane technology, in convenient spin-column or 96-well formats, ensures fast and reproducible DNA purification, eliminating the need for organic extraction and alcohol precipitation (see DNeasy Mini and 96 procedures). Samples are first lysed using proteinase K. Buffering conditions are adjusted to provide optimal DNA-binding conditions and the lysate is loaded onto the DNeasy Mini spin column or the DNeasy 96 plate. During centrifugation, DNA is selectively bound to the DNeasy membrane as contaminants pass through. Remaining contaminants and enzyme inhibitors are removed in two efficient wash steps and DNA is then eluted in water or buffer, ready for use.
DNeasy Mini spin columns are prepackaged in collection tubes and individually sealed, providing convenience and safety. The purification procedure using DNeasy Mini spin columns can be automated on the QIAcube. The DNeasy 96 Blood & Tissue Kit provides high-throughput processing in a 96-well format using the QIAGEN 96-Well-Plate Centrifugation System.
See figures
Applications
DNeasy Blood & Tissue Kits provide high-quality DNA, ready to use in all downstream assays, including applications in:
- Life science research
- Livestock breeding
- Pedigree genotyping
- Veterinary pathogen research
- Routine applied testing
Supporting data and figures
DNeasy Blood & Tissue Kit (250) ACT environmental impact factor label US.
ACT environmental impact factor labels are designed to evaluate and score products on several sustainability criteria. Products are score 1-10 except for energy and water consumption, which are scored as 1 point per kWh or gallon, respectively. A low score means a lower environmental impact.

Specifications
| Features | Specifications |
|---|---|
| Applications | PCR, real-time PCR, genotyping |
| Elution volume | 100–200 µl |
| Time per run or per prep | 20 minutes – 1 hour |
| Main sample type | Blood, tissue |
| Format | 96-well plate, spin column |
| Sample amount | 100 µl/25 mg |
| Processing | Manual |
| Yield | 6 µg/30 µg |
| Technology | Silica technology |
| Purification of total RNA, miRNA, poly A+ mRNA, DNA or protein | DNA |