✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
EZ2 RNA/miRNA Tissue/Cells Kit (48)
Cat. No. / ID: 959035
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Purifies RNA and miRNA from human and animal cells and tissues
- Does not use toxic phenol
- Includes highly efficient enzymatic removal of protein and gDNA
- Lets you process up to 24 samples at the same time, for easy intra-run comparability
- Prevents RNase contamination with prefilled cartridges and closed workdeck
Product Details
The EZ2 RNA/miRNA Tissue/Cells Kit makes it easy and safe to purify total RNA – including miRNA and other small RNA – from cultured cells and various human or animal tissues. The process requires no use of phenols, and once the lysis step is completed, all remaining steps are automated with any EZ2 Connect instrument.
Performance
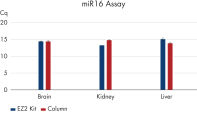
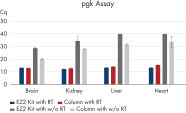
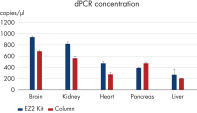
The EZ2 RNA/miRNA Tissue/Cells protocol isolates a similar amount of total RNA, including small RNA, to manual column-based protocols. This is true even for difficult tissue types such as fibrous tissue (e.g., muscle) or fatty tissue (e.g., brain) (see Comparable miRNA recovery). Genomic DNA removal is likewise comparable to manual column-based purification (see Efficient mRNA recovery and gDNA removal). Digital PCR analysis shows higher RNA concentration in samples processed with the EZ2 RNA/miRNA Tissue/Cells Kit (see Higher RNA concentration).
See figures
Principle
The EZ2 RNA/miRNA Tissue/Cells protocol uses manual lysis in Buffer RLT protect the RNA molecules. Proteinase K digestion ensures complete lysis of even hard-to-lyse tissues. Bind, wash, and elute steps are automated on an EZ2 Connect instrument, which uses magnetic bead technology. After first binding, RNA is treated with DNase to digest contaminating genomic DNA, and then rebound to the magnetic beads. Wash steps then remove contaminants that could interfere with enzymatic reactions that follow.
Procedure
The cell pellet or cell culture is first homogenized, or the tissue sample is disrupted and then homogenized. RNase-free water and Proteinase K are then added, and the mixture is incubated for 10 minutes at room temperature (see EZ2 RNA/miRNA Tissue/Cells workflow).
After incubation, samples and prefilled reagent cartridges are loaded into the EZ2 Connect, and application run is started. Once the run is finished, purified RNA or miRNA will be ready for downstream applications (see EZ2 RNA/miRNA Tissue/Cells workflow).
See figures
Applications
RNA and miRNA purified with the EZ2 RNA/miRNA Tissue/Cells Kit is highly suitable for RT-PCR, digital PCR (dPCR) or next-generation sequencing (NGS).
Supporting data and figures
EZ2 RNA/miRNA Tissue/Cells workflow.

Specifications
| Features | Specifications |
|---|---|
| Applications | RT-PCR, dPCR, NGS |
| Elution volume | 50 or 100 µl |
| Main sample type | Cells, tissue |
| Processing | Automated with an EZ2 Connect instrument |
| Analyte | Total RNA, including miRNA and mRNA |
| Sample amount | Up to 30 mg (frozen) or 15 mg (stabilized) tissue or 10 mg frozen spleen or lung tissue, or up to 5 x 10^6 cells |
| Technology | Magnetic-particle technology |