QIAseq Targeted RNA Custom Panel (96)
Cat. No. / ID: 333025
Features
- Choose from comprehensive panels or select up to 1000 genes and build a custom panel
- UMIs (unique molecular indices) ensure accurate gene expression profiling
- Suitable as an alternative to qPCR for gene expression with >384 genes in more than 24 samples
- Includes access to GeneGlobe data analysis portal for read alignment, data normalization and gene expression analysis
- Available for Illumina and Thermo-Fisher NGS instruments
Product Details
QIAseq Targeted RNA Panels have been developed as a Sample to Insight solution for quantitative gene expression profiling using RNA-seq. These panels integrate unique molecular indices (UMIs) and single primer extension (SPE) in a simple PCR-based library preparation workflow to deliver unbiased and accurate quantification from human, mouse and rat samples.
QIAseq Targeted RNA Indices are for indexing samples for targeted RNA sequencing and primers necessary for sequencing RNA libraries generated by QIAseq Targeted RNA Panels.
Performance
- Accuracy: Innovative UMIs (molecular barcode counting) eliminate PCR duplication and amplification bias to deliver the most accurate results (see figure "Unbiased and accurate gene quantification").
- Specificity: The unique combination of our proprietary primer design algorithm and rigorous testing of every primer assay guarantees high specificity and accurate results (see figure "Proprietary primer design delivers gene-specific amplicons – 97% specificity").
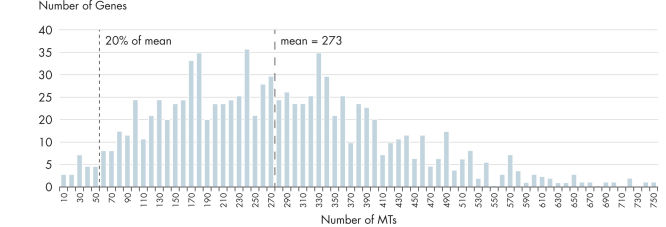
- Uniformity: The QIAseq Targeted RNA Panel workflow has been optimized to deliver highly uniform sequencing results, to ensure sequencing capacity is utilized very efficiently. In fact, UMIs (unique molecular indices) entirely remove PCR bias in RNA-seq counting (see figure "Unmatched uniformity – 97% of assays are within 20% of median molecular tag counts").
- Reproducibility: The QIAseq Targeted RNA Panel system demonstrates strong correlations across technical replicates, product lots and instruments with correlation coefficients averaging above >0.99, to ensure reliable detection of differences in expression between biological samples.
- Sensitivity: Targeted RNA sequencing using UMIs is optimized to deliver highly reliable quantification down to ~100 copies of an RNA target in 25 ng total RNA (see figure "Positive results with as little as 0.2 copies of RNA per cell"). Low-input protocols allow researchers to start with lower amounts of RNA in smaller volumes.
- Flexibility: QIAseq panels combine the power of NGS with the accuracy of qPCR to allow multiplexing of several samples per NGS while delivering cost-effective results (see figure "Simple procedure").
Principle
Traditional RNA sequencing methods suffer from PCR duplication and amplification bias, resulting in inaccurate gene expression analysis. By introducing UMIs (unique molecular indexes) before any amplification takes place, QIAseq Targeted RNA Panels are able to eliminate this issue to deliver accurate and digital quantification of genes (see figure "Unbiased and accurate gene quantification").
A unique feature of the QIAseq Targeted RNA Panels is the set of built-in control assays. The gDNA assays control for any gDNA contamination in the RNA sample to ensure reproducible results. The housekeeping gene (HKG) assays are used to normalize data, thereby making sample-to-sample and run-to-run comparisons possible.
Procedure
The QIAseq Targeted RNA Panel workflow begins with converting total RNA into cDNA (see figure "Simple procedure"). The workflow requires minimal RNA input: typically 25 ng of total RNA can be used. No enrichment or rRNA depletion steps are necessary. The molecular indexing step makes use of a gene-specific primer (GSP1) which contains a 12-base UMI in a multiplex primer panel (targeting 12–1000 genes). After the barcoding step, the uniquely tagged cDNA is bead purified to remove residual primers, and a PCR is set up with a second pool of gene-specific adapter primers (GSP2) and the RS2 primer, which primes off of a common tag on the GSP1 primers. This reaction insures that intended targets are enriched sufficiently to be represented in the final library. The number of cycles is kept to a minimum to keep PCR-induced variations in amplification to a low level (any variations are easily corrected and accounted for with the molecular barcodes). Another quick cleanup with beads is performed, and a universal PCR is run with RS2 and FS2 primers, which also adds sample-indexing barcodes to each sample. A final cleanup with beads is performed and the library is complete, and ready for quantification and sequencing.
Purchase of QIAseq targeted RNA Panels provides access to data analysis tools at QIAGEN’s GeneGlobe portal. These include read alignment and data normalization and differential gene expression at no additional costs.
Applications
- Gene expression profiling
- Biomarker research
- Confirmation of whole transcriptome sequencing data
- Confirmation of microarray data
Supporting data and figures
Unmatched uniformity (>97% of assays are within 20% of median molecular tag counts).
A 917-plex gene panel was used to prepare a library from 10 ng of NA12878 reference DNA. All assays were designed to be intra-exon, and are thus single copy on genomic DNA. This allows an estimation of the uniformity of amplicon performance in the library preparation step (e.g., every unique tag equals one captured copy). In terms of raw assay performance, 97.5% of assays are within 20% of mean/median molecular tag counts. For cataloged panels, any assay below 20% is redesigned and replaced. UMIs (unique molecular indexes) entirely remove this variation in RNA-seq counting.