Rotor-Gene Multiplex RT-PCR Kit
For ultrafast, multiplex, one-step qRT-PCR gene expression analysis on Rotor-Gene cyclers
For ultrafast, multiplex, one-step qRT-PCR gene expression analysis on Rotor-Gene cyclers
The Rotor-Gene Multiplex RT-PCR Kit is designed for use with the Rotor-Gene Q and other Rotor-Gene cyclers, providing ultrafast, highly reliable quantification in multiplex, real-time one-step RT-PCR using sequence-specific probes. Depending on the cycler configuration, up to 4 RNA targets (e.g., 1 control gene and 3 target genes) can be quantified simultaneously in the same tube. Outstanding performance is achieved through the combination of a specially optimized master mix and the unique Rotor-Gene cycler. For convenience, the master mix can be stored at 2–8°C.
IMPORTANT NOTE: As announced earlier, the production of the Rotor-Gene kits has been discontinued since mid-2021. Hence, these products will be available only until stocks last. Visit the product page of the successor kit to view improved features or to request a trial kit.
For more information and FAQs on this transition, visit: www.qiagen.com/PCRresource.
Amplifying reference and target genes in the same reaction instead of in separate reactions increases the reliability of gene quantification by minimizing handling errors. The Rotor-Gene Multiplex RT-PCR Kit enables reliable multiplex quantification of RNA targets on the Rotor-Gene Q without the need for optimization of reaction and cycling conditions (see flowchart " QIAGEN multiplex kits"). Real-time one-step RT-PCR is carried out, enabling reverse transcription and PCR to take place sequentially in the same reaction vessel. Since it is not necessary to transfer the finished reverse transcription reaction to another tube for PCR, the real-time RT-PCR procedure is streamlined, making high-throughput analysis possible.
Highly specific amplification is assured through a balanced combination of K+ and NH4+ ions, which promote specific primer annealing and enable high PCR specificity and sensitivity, while synthetic Factor MP, an innovative PCR additive specially developed for challenging multiplex PCR applications, allows different amplicons in the same reaction to all be amplified with the same high efficiency (see figure " Unique PCR buffer").
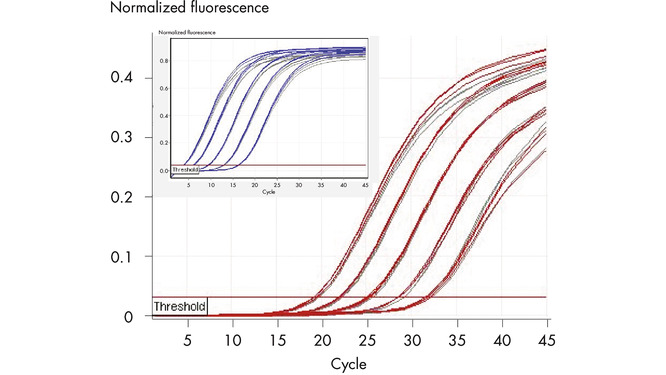
Fast cycling without compromising performance is achieved using Q-Bond, a novel PCR additive that considerably shortens cycler run times (see figure " Fast primer annealing"). In addition, an optimized mix of reverse transcriptases provides efficient cDNA synthesis in just 15 minutes, while the highly stringent hot-start enzyme HotStarTaq Plus DNA Polymerase is rapidly activated at the start of PCR by a brief 5-minute incubation at 95ºC.
| Component | Features | Benefits |
|---|---|---|
| HotStarTaq Plus DNA Polymerase | 5 min activation at 95ºC | Set up of qPCR reactions at room temperature |
| Rotor-Gene Multiplex RT-PCR Buffer | Balanced combination of NH4+ and K+ ions | Specific primer annealing ensures reliable qPCR results |
| Synthetic Factor MP | Reliable multiplexing analysis of up to 4 genes in the same tube | |
| Unique Q-Bond additive | Faster PCR run times enable faster results and more reactions per day | |
| Rotor-Gene RT Mix | Special blend of reverse transcriptases with a high affinity for RNA | RNA can be transcribed in just 15 minutes, even through complex secondary structures |
A ready-to-use master mix eliminates the need for optimization of reaction and cycling conditions, such as primer and probe concentrations. Simply add template RNA, primer–probe sets, and the supplied reverse transcriptase mix to the master mix and program the cycler. The handbook supplied with the kit lists recommended dyes and contains a single protocol for all multiplex RT-PCR assays.
The Rotor-Gene Multiplex RT-PCR Kit is optimized for fast, real-time one-step RT-PCR analysis using sequence-specific probes on the Rotor-Gene Q. It is also compatible with the Rotor-Gene 3000 and the Rotor-Gene 6000. Up to 4 RNA targets can be simultaneously and rapidly quantified in the same tube, increasing throughput and saving precious sample material.
| Features | Specifications |
|---|---|
| Applications | Real-time quantification of RNA targets in a multiplex format |
| Real-time or endpoint | Real-time |
| Single or multiplex | Multiplex |
| With or without ROX | Without ROX dye |
| SYBR Green I or sequence-specific probes | Sequence-specific probes |
| Reaction type | Real-time one-step RT-PCR |
| Sample/target type | RNA |
| Thermal cycler | Rotor-Gene Q, Rotor-Gene 3000, Rotor-Gene 6000 |