miRCURY LNA Premium miRNA Mimic (5 nmol)
Cat. No. / ID: 339178
Features
- Third generation of highly potent, mature miRNA mimics with unique triple RNA strand design
- No miRNA-star activity from bisected LNA-enhanced passenger strand
- miRNA strand sequence matches miRBase annotation
- Available with fluorescent label to assess transfection efficiency
- Available with biotinylated miRNA strand to isolate targets by RNA pull-down
Product Details
miRCURY LNA miRNA Mimics are designed to simulate naturally occurring mature miRNAs. Introducing an miRNA mimic into cells increases the proportion of RNA-induced silencing complexes (RISC) containing the guide strand miRNA, enabling assessment of the phenotypic consequences of increased activity and discovery of new miRNA functions. miRCURY LNA miRNA Mimics have an innovative design that includes two short, LNA-enhanced complimentary strands that prevent any miRNA-like activity associated with the passenger strands, so you can be sure that phenotypes observed using these mimics are due to increased activity of the mimicked miRNA.
Need a quote for your research project or would you like to discuss your project with our specialist team? Just contact us!
Performance
Excellent potency
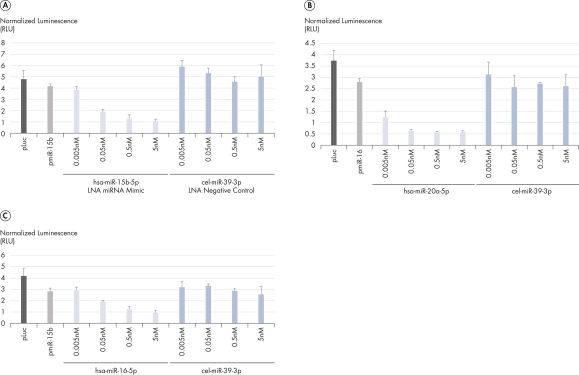
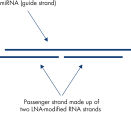
The miRNA strand of our mimics is made of unmodified RNA to provide as accurate miRNA mimicry as possible. The sequence and LNA spiking pattern of the two complementary passenger strands have been carefully designed to optimize efficient incorporation of the miRNA (guide) strand into RISC (see figure Silencing of miRNA reporters using mimics for examples of the high potency of these LNA mimics).No miRNA star-like activity from the passenger strand
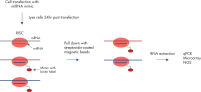
miRCURY LNA miRNA Mimics have an innovative design, which includes two short LNA-enhanced complimentary strands that prevent any miRNA-like activity associated with the passenger strands (see figure The distinct RNA triplex design ensures completely specific miRNA mimicry). This guarantees that phenotypes observed using these mimics are due to increased activity of the mimicked miRNA.Fluorescently labeled miRNA mimics
Fluorescently labeled miRNA mimics can be useful to assess transfection efficiency. However, addition of fluorescent labels to the 5’ or 3’ end of the miRNA strand completely removes activity of miRNA mimics. In contrast, fluorophores at the 5’ end of the passenger strand are tolerated and have no adverse effects on the resulting mRNA activity (see figures Fluorescently labeled miRNA mimics and Biotinylated miRCURY LNA miRNA mimics). Because the fluorophore is attached to the passenger strand, the fluorescent signal does not report on the intracellular localization of the miRNA strand that is loaded into RISC and is only useful for monitoring transfection efficiency (see figure Silencing of miRNA reporter assays with fluorescently labeled miRCURY LNA miRNA Mimics).Biotinylated miRNA mimics
Modification of the 5’ and 3’ ends of the miRNA strand in mimics is generally not well tolerated at all. However, we have observed that potent miRNA activity is achievable with biotinylated mimics, although they are significantly less potent than non-modified mimics, presumably due to less efficient loading into RISC. Nevertheless, the possibility that biotin modification of the guide strand creates a bias for incorporation of the passenger strand cannot be excluded. It is therefore important to use modified mimics that ensure that only the miRNA strand is chosen by RISC. The special triple RNA strand design of miRCURY LNA miRNA mimics ensures complete miRNA strand specificity, since the passenger strand is split into two LNA-modified RNA strands (see figures The distinct RNA triplex design ensures completely specific miRNA mimicry and Silencing of miRNA reporter assays with biotinylated miRCURY LNA miRNA Mimics).See figures
 The distinct RNA triplex design ensures completely specific miRNA mimicry.
The distinct RNA triplex design ensures completely specific miRNA mimicry.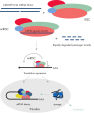 Fluorescently labeled miRNA mimics.
Fluorescently labeled miRNA mimics.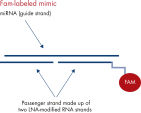 Biotinylated miRCURY LNA miRNA Mimics.
Biotinylated miRCURY LNA miRNA Mimics.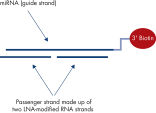 Silencing of miRNA reporter assays with fluorescently labeled miRCURY LNA miRNA Mimics.
Silencing of miRNA reporter assays with fluorescently labeled miRCURY LNA miRNA Mimics. Silencing of miRNA reporter assays with biotinylated miRCURY LNA miRNA Mimics.
Silencing of miRNA reporter assays with biotinylated miRCURY LNA miRNA Mimics.
Principle
miRCURY LNA miRNA Mimics have a unique and innovative design that includes three RNA strands, rather than the two RNA strands that characterize traditional miRNA mimics. The miRNA (guide) strand is an unmodified RNA strand with a sequence corresponding exactly to the annotation in miRBase. However, the passenger strand is divided into two LNA-enhanced RNA strands (see figure A unique triple RNA strand design). When designed correctly, these triple RNA strand mimics are as potent as traditional double-strand RNA mimics. The great advantage is that the segmented nature of the passenger strand ensures that only the miRNA strand is loaded into the RNA-induced silencing complex (RISC) with no resulting miRNA activity from the two complementary passenger strands. Phenotypic changes observed with miRCURY LNA miRNA mimics can therefore be safely ascribed to the miRNA simulated by the mimic (see figure miRNA target identification with biotinylated mimics).
The distinct triple RNA strand design is enabled by incorporation of high-affinity LNA nucleotides into the two passenger strands. The sequence, length and LNA spiking pattern of the two passenger strands have been optimized using a sophisticated and empirically derived design algorithm.
miRCURY LNA miRNA Mimics have been predesigned for a large majority of human, mouse and rat miRNAs listed in miRBase. Since many miRNAs are phylogenetically conserved, this set of miRNA mimics covers a large proportion of vertebrate and invertebrate miRNAs. >
See figures
Procedure
Applications
miRNA mimics are also frequently used for validating miRNA targets in combination with miRNA inhibitors and target site blockers. Typically, plasmid-based assays are used in which the 3' UTR of the mRNA under investigation has been cloned downstream of a reporter gene. Introducing the mimic into cells harboring the reporter plasmid will reduce reporter gene expression, while miRNA inhibitors and target site blockers masking the 3' UTR miRNA binding site will cause derepression.
Biotinylated miRNA mimics are highly effective tools for identification of miRNA targets in RNA pull-down experiments. Recent advances with this experimental approach are transforming our understanding of miRNA biology and have revealed that non-canonical miRNA–mRNA interactions, which are ignored by target prediction tools, are frequent and lead to target repression. The principle of the technique is shown in Perfect miRNA strand-specific activity with miRNA mimics. Biotinylated mimics are significantly less potent than non-modified mimics.
Supporting data and figures
Silencing of miRNA reporter assays with miRCURY LNA miRNA Mimics.