EpiTect Hi-C Kit (US)
A single-box solution for high-resolution mapping of chromatin folding
A single-box solution for high-resolution mapping of chromatin folding
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Cat. No. / ID: 59971
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Hi-C technology is a powerful technique for genome-wide chromosome conformation capture that enables the characterization of chromatin folding at kb resolution.
The EpiTect Hi-C Kit offers a robust, yet simple and fast, protocol with low cell input requirements that enables generation of high-quality Hi-C Illumina NGS libraries from cross-linked cells in less than 2 days.
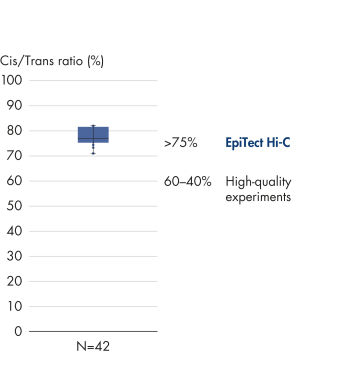
The EpiTect Hi-C Kit generates high-quality Hi-C NGS libraries, ensuring that first-rate data is generated from costly downstream deep sequencing. Sequencing results from more than 40 EpiTect Hi-C libraries have been analyzed to evaluate the performance of the kit. The most important QC metrics are shown in the following figures: Percentage of Hi-C events, Percentage of long-range cis interactions, Cis/Trans ratio, No strand orientation bias with the EpiTect Hi-C Kit and Percentage of paired reads deriving from a single restriction fragment. The data show that the EpiTect Hi-C Kit generates NGS libraries that, on average, far exceed criteria normally considered sufficient for a successful Hi-C experiment.
Hi-C is a proximity ligation assay that captures chromatin interactions on a genome-wide scale. The EpiTect Hi-C Kit is a specialized DNA preparation resulting in an Illumina-compatible NGS library (see figures EpiTect Hi-C workflow – day 1 and EpiTect Hi-C workflow – day 2). Briefly described, the assay starts with the purification of nuclei in which chromatin conformation has been frozen by chemical cross-linking of DNA binding proteins and DNA. The DNA is then completely digested with a 4 bp restriction enzyme. Open DNA ends are labeled with biotin and subsequently ligated. Paired-end sequencing of the Hi-C ligation products identifies very large numbers of chimeric sequences that derive from DNA strands that were closely associated in space. The probability that two sequences are ligated together is a function of their average distance in space. Quantification of ligation junctions allows for the determination of DNA contact frequencies from which high-resolution mapping of chromatin folding can be achieved.
The EpiTect Hi-C workflow (see figures EpiTect Hi-C workflow – day 1 and EpiTect Hi-C workflow – day 2) represents a marked improvement over published protocols. A week-long and complicated procedure has been converted into a simple and robust protocol that requires just 1.5 days. Furthermore, the sample input requirement has been reduced by one order of magnitude, allowing creation of Hi-C NGS libraries from just 5000 cells. The protocol has been developed for work with cross-linked cells from mammalian cell cultures.
The EpiTect Hi-C procedure is a version of the in situ (i.e., in nucleus) Hi-C method in which nuclei are gently purified and permeabilized to maintain the spatial organization of the genome during the initial digestion and ligation steps. This process is vital in order to minimize background noise from uninformative ligation events that do not reflect genome organization. This is because intact nuclei constrain the movement and random collisions of cross-linked complexes, such that ligation events predominantly occur between topologically associated DNA fragments.
Constructing Hi-C NGS libraries
The EpiTect Hi-C Kit workflow consists of 2 parts and each can be completed in one day. The steps of the protocol are summarized in the tables below and visualized in figures EpiTect Hi-C workflow – day 1 and EpiTect Hi-C workflow – day 2. The included Illumina adapters have sequence bar codes that enable multiplex sequencing of up to 6 samples.
To view the full protocol, see our detailed EpiTect Hi-C Handbook.
Data analysis
Hi-C data analysis is offered at our GeneGlobe Data Analysis Center. Hi-C sequencing results can be analyzed using the EpiTect Hi-C Data Analysis Portal, which uses open-source tools to provide a QC sequencing report, Hi-C contact matrices and visualization of chromatin contact maps. For more information see our EpiTect Hi-C Data Analysis Portal User Guide.
Phase Genomics, Inc, offers a comprehensive suite of computational analytic services for Hi-C data analysis using both open source as well as proprietary software. Services include data QC, Hi-C matrix calculation and visualization, identification of architectural features such as A/B compartments, TADs and loops, structural variation analysis, genome assembly and haplotype phasing, and other custom analyses. A more detailed description of analytic options can be found at phasegenomics.com and a list of publications can be found at Publications & News.
Chromatin conformation
The three-dimensional organization of chromatin is under intense investigation because it has a profound influence on genome function. By enabling the capture of long-range DNA contacts on a genome-wide scale, Hi-C has quickly become a very important tool for the analysis of nuclear organization. Analysis of Hi-C data has revealed the amazing complexity of genome architecture, with multiple layers of spatial organization that partition the genome into chromosome territories, chromosomal sub-compartments, topologically associated domains (TADs) and DNA loops at increasing resolution (see figure Levels of chromatin organization). In addition, genome organization is dynamic and changes during development. In no two cell types do chromosomes fold alike.
Hi-C technology is refining our understanding of the underlying mechanisms of gene regulation by demonstrating the crucial role played by chromatin folding. For example, Hi-C data have provided overwhelming, concrete experimental evidence for the long-held belief that enhancers and cognate promoters are joined together in space by DNA loops, explaining how regulatory elements can exert control of gene expression over great distances of genome sequence. In addition, Hi-C data have revealed that chromatin folding helps establish separate nuclear compartments with distinct regulatory environments by grouping together, in space, DNA domains that are distal in sequence, but which share the same set of epigenetic marks. Importantly Hi-C data is also revealing how mutations that cause subtle changes in chromatin structure can alter gene expression dramatically and cause disease.