用于在Rotor-Gene PCR仪上超快速的多重一步法qRT-PCR基因表达分析
Features
- 在Rotor-Gene PCR仪上获得超快速、可靠的结果
- 单管内高灵敏检测多个RNA靶序列
- 表现优越的多重PCR,无需优化
- 低丰度和高丰度靶基因高效共扩增
Product Details
Rotor-Gene Multiplex RT-PCR Kit 专用于Rotor-Gene Q实时荧光定量PCR分析仪和其他Rotor-Gene PCR仪,可超快速进行基于序列特异性探针的可靠的多重一步法real-time RT-PCR定量分析。通过调整PCR仪,可在单管内同步定量分析多至4个RNA靶基因(如1个参照和3个靶基因)。配合经优化的预混液和具有独特转子设计的Rotor-Gene PCR仪共同使用,表现卓越。为便于使用,预混液应储存在2–8°C。
Performance
See figures
Principle
在同一反应管中扩增参照和靶基因,减少了手动操作失误,提高了基因定量检测的可靠性。Rotor-Gene Multiplex RT-PCR Kit能够在Rotor-Gene Q实时荧光定量PCR分析仪上可靠的多重定量检测RNA靶基因,无需优化反应条件(参见" QIAGEN multiplex kits")。进行一步法real-time RT-PCR,使逆转录和PCR扩增在同一反应容器中相继进行。因为无需将逆转录产物转移到另一管中进行PCR,所以real-time RT-PCR的流程得以简化,可实现高通量分析。
平衡的K+和NH4+离子组合确保高度特异性,促进特异性引物退火,PCR特异性高、灵敏度好。一种新颖的PCR添加剂合成的Factor MP专用于多重PCR应用,能够以同样的效率扩增在同一反应中不同的扩增子(参见" Unique PCR buffer")。
一种新颖的PCR添加剂Q-Bond可缩短扩增时间,确保快速扩增的同时不影响表现(参见" Fast primer annealing")。此外,逆转录酶在15分钟内即可高效的合成cDNA,而高度严谨的热启动酶HotStarTaq Plus DNA Polymerase在95ºC条件下只需5分钟即可快速激活。
| 组分 | 特点 | 优势 |
|---|---|---|
| HotStarTaq Plus DNA Polymerase | 95ºC条件下5分钟激活 | 室温下建立qPCR反应体系 |
| Rotor-Gene Multiplex RT-PCR Buffer | 浓度平衡的NH4+和K+离子 | 特异性引物退火,确保可靠的qPCR结果 |
| 合成的Factor MP | 在同一反应管中可靠的进行多达4个基因的多重分析 | |
| 独特的Q-Bond添加剂 | 更快速的PCR循环,快速获得结果,一天可进行更多的反应 | |
| Rotor-Gene RT Mix | RNA亲和性高的逆转录酶 | RNA逆转录可在15分钟内完成,甚至包括复杂的二级结构 |
See figures
Procedure
应用即用型预混液无需优化反应和循环条件。只需在预混液中添加模板RNA和引物探针对、逆转录酶,然后设置PCR仪即可开始实验。试剂盒中提供的操作手册中列出了推荐染料,并为多重RT-PCR分析提供了单独的实验方案。
Applications
Supporting data and figures
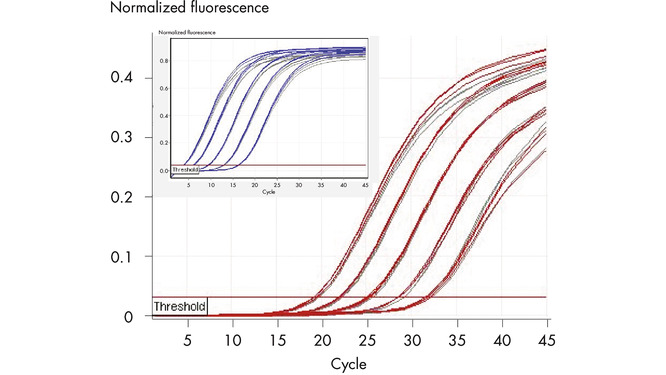
Reliable duplex analysis.
Specifications
| Features | Specifications |
|---|---|
| Applications | Real-time quantification of RNA targets in a multiplex format |
| Real-time or endpoint | Real-time |
| Single or multiplex | Multiplex |
| With or without ROX | Without ROX dye |
| SYBR Green I or sequence-specific probes | Sequence-specific probes |
| Reaction type | Real-time one-step RT-PCR |
| Sample/target type | RNA |
| Thermal cycler | Rotor-Gene Q, Rotor-Gene 3000, Rotor-Gene 6000 |






