EpiTect Bisulfite Kits
亚硫酸氢盐的完全转化和DNA回收,用于甲基化分析
亚硫酸氢盐的完全转化和DNA回收,用于甲基化分析
✓ 全天候自动处理在线订单
✓ 博学专业的产品和技术支持
✓ 快速可靠的(再)订购
Cat. No. / ID: 59104
✓ 全天候自动处理在线订单
✓ 博学专业的产品和技术支持
✓ 快速可靠的(再)订购
完整的EpiTect离心和真空过程只需6小时,包括从DNA到待分析的亚硫酸氢盐转化后的高纯DNA。EpiTect 96 Bisulfite操作时间少于7小时。传统的亚硫酸氢盐转化过程需要18个小时,且需要大量的操作(参见" 快速简单的亚硫酸氢盐转化"和" Time saving procedure")。EpiTect Bisulfite Kit为成功进行亚硫酸氢盐转化和DNA回收提供所需的全部试剂。
| EpiTect流程 | 传统流程 | |
|---|---|---|
| 反应构建 | 10分钟;预制溶液 | 40分钟;需反应构建 |
| 亚硫酸氢盐转化 | 5小时 | 16小时 |
| 纯化 | 30–45分钟; EpiTect离心柱或孔板 | 120分钟;纯化、脱璜、沉淀和洗涤 |
每个EpiTect Bisulfite Kit可以相同的效率转化低至1 ng到2 µg DNA。全新的实验步骤和优化的缓冲液可回收得到高产量的转化DNA。当通过real-time PCR扩增经转化的DNA时,低CT值的实验结果显示了高效的胞嘧啶转化(参见" Complete cytosine conversion")。对转化的DNA的分析显示,EpiTect Bisulfite Kit可转化超过99%的非甲基化的胞嘧啶(参见" Highly efficient cytosine conversion")。高转化率确保了高重复性和可靠的下游分析结果。
从珍稀样本和少量样本(如:福尔马林固定、FFPE石蜡包埋或显微切割组织)中确定甲基化模式,由于DNA的量很少,实验尤其困难。但是这些样本类型是成功鉴定有价值的疾病,预测生物标记物的专门研究对象。 EpiTect Bisulfite Kit 为这些类型的样本提供优化的操作流程,从而解决了难题(参见" Methylation detection from FFPE tissue samples")。该试剂盒含有实验所需的全部缓冲液和试剂,只需少量的手动操作。
对于从FFPE样本、细胞、血液及组织中直接进行亚硫酸氢盐转化而无需预先分离DNA,我们推荐使用EpiTect Plus FFPE Bisulfite Kit和EpiTect Plus LyseAll Bisulfite Kit。
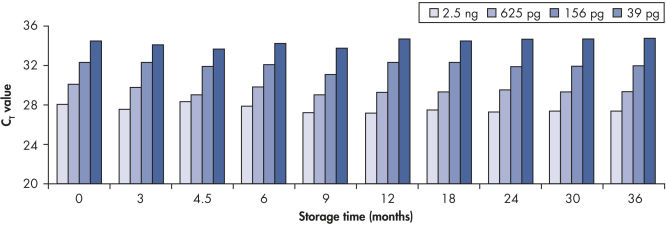
DNA保护缓冲液采用独特的配方,防止DNA过度降解(在高温、低pH值条件下使用亚硫酸氢盐处理DNA时经常出现严重的DNA降解),并实现有效的DNA变性,从而产生完全转化为胞嘧啶时所需的单链DNA(参见" Unique DNA Protect Buffer and pH indicator system")。防止DNA降解的发生可以保证后续大PCR片段的扩增和分析(参见" Fast and easy bisulfite conversion")。DNA保护缓冲液含有PH指示剂染料,从而可保证用于胞嘧啶转化的正确的pH条件。有效的完整DNA回收可保证转化后的DNA至少保存12个月而不影响DNA的质量(参见" Long-term DNA storage")。
EpiTect Bisulfite操作流程基于QIAGEN简单直接的结合、洗涤和洗脱体系,仅需几个简单的步骤(参见" Conversion procedure")。DNA经过热变性和亚硫酸氢盐转化后,被上样到EpiTect离心柱或96孔板上,使用优化的缓冲液和标准化的离心步骤或真空装置,洗去所有的亚硫酸氢钠并洗脱DNA。洗脱的DNA可用于各种下游应用,如:甲基特异性PCR、real-time PCR分析、亚硫酸氢盐转化后测序(bisulfite sequencing)、结合亚硫酸氢盐的限制性内切酶法分析(COBRA)和焦磷酸测序(Pyrosequencing)。
表观遗传学信息对于生物医学研究(尤其是肿瘤学)、干细胞研究以及发育生物学等领域都非常重要。但是分析DNA甲基化的变化目前仍具有挑战性,因为缺乏标准化的方法可从有限的样本中得到可重现性的数据。QIAGEN推出新的表观遗传学解决方案,使样本分析前处理到分析整个过程实现标准化,即从DNA样本收集、稳定和纯化,到亚硫酸氢盐转化和real-time或终点式PCR甲基化分析或测序(参见" Standardized workflows in epigenetics")。
经EpiTect Bisulfite Kit转化和纯化的DNA适用于多种下游应用,如:
Bisulfite converted DNA generated with EpiTect Kits was stored at –20°C for up to 36 months, and amplified at several time intervals. The resulting threshold cycle values at all time points for various DNA amounts were comparable to those obtained with freshly converted DNA (time point 0).
| Features | Specifications |
|---|---|
| Applications | Multiplex PCR, real-time PCR, sequencing |
| Processing | Manual |
| Sample types | Blood, (FFPE) tissue, DNA samples |
| Conversion efficiency | 99.4–99.8% |
| Format | Spin column and 96-well plate |
| Time per run or per prep | <6 hours (1-48 samples), ~ 7 hours (48-96 samples) |
| Elution volume | Varies |
| Sample amount | 1–2 µg |
| Yield | Depends on input amount of purified genomic DNA used for conversion |